G3: Inexpensive GBS, new algorithm for genetic mapping
Rapid and Inexpensive Whole-Genome Genotyping-by-Sequencing for Crossover Localization and Fine-Scale Genetic Mapping
Beth A. Rowan, Vipul Patel, Detlef Weigel, and Korbinian Schneeberger
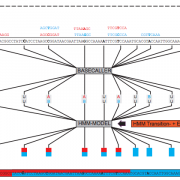
The reshuffling of existing genetic variation during meiosis is important both during evolution and in breeding. The re-assortment of genetic variants relies on the formation of crossovers (COs) between homologous chromosomes. The pattern of genome-wide CO distributions can be rapidly and precisely established by short read sequencing of individuals from F2 populations, which in turn are useful for quantitative trait locus (QTL) mapping. While sequencing costs have dropped precipitously in recent years, the costs of library preparation for hundreds of individuals have remained high. To enable rapid and inexpensive CO detection and QTL mapping using low-coverage whole-genome sequencing of large mapping populations, we have developed a new method for library preparation along with Trained Individual GenomE Reconstruction (TIGER), a probabilistic method for genotype and CO predictions for recombinant individuals. In an example case with hundreds of F2 individuals from two Arabidopsis thaliana accessions, we resolved most CO breakpoints to within 2 kb and reduced a major flowering time QTL to a 9-kb interval. In addition, an extended region of unusually low recombination revealed a 1.8-Mb inversion polymorphism on the long arm of chromosome 4. We observed no significant differences in the frequency and distribution of COs between F2 individuals with and without a functional copy of the DNA helicase gene RECQ4A. In summary, we present a new, cost efficient method for large-scale, high-precision genotyping-by-sequencing.