On bioRxiv: Arabidopsis genome assembled with one Nanopore flowcell
High contiguity Arabidopsis thaliana genome assembly with a single nanopore flow cell
Arabidopsis thaliana genome assembly with a single Oxford Nanopore flow cell
Todd P. Michael, Florian Jupe, Felix Bemm, Stanley T. Motley, Justin P. Sandoval, Olivier Loudet, Detlef Weigel, VJoseph R. Ecker
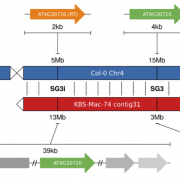
While many evolutionary questions can be answered by short read re-sequencing, presence/absence polymorphisms of genes and/or transposons have been largely ignored in large-scale intraspecific evolutionary studies. To enable the rigorous analysis of such variants, multiple high quality and contiguous genome assemblies are essential. Similarly, while genome assemblies based on short reads have made genomics accessible for non-reference species, these assemblies have limitations due to low contiguity. Long-read sequencers and long-read technologies have ushered in a new era of genome sequencing where the lengths of reads exceed those of most repeats. However, because these technologies are not only costly, but also time and compute intensive, it has been unclear how scalable they are. Here we demonstrate a fast and cost effective reference assembly for an Arabidopsis thaliana accession using the USB-sized Oxford Nanopore MinION sequencer and typical consumer computing hardware (4 Cores, 16Gb RAM). We assemble the accession KBS-Mac-74 into 62 contigs with an N50 length of 12.3 Mb covering 100% (119 Mb) of the non-repetitive genome. We demonstrate that the polished KBS-Mac-74 assembly is highly contiguous with BioNano optical genome maps, and of high per-base quality against a likewise polished Pacific Biosciences long-read assembly. The approach we implemented took a total of four days at a cost of less than 1,000 USD for sequencing consumables including instrument depreciation.