In Nature Plants: Prominent TADs in rice
Prominent topologically associated domains differentiate global chromatin packing in rice from Arabidopsis
Chang Liu, Ying-Juan Cheng, Jia-Wei Wang & Detlef Weigel
Nature Plants (2017) doi.org/10.1038/s41477-017-0005-9
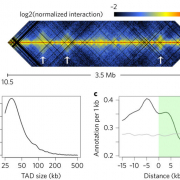
The non-random three-dimensional organization of genomes is critical for many cellular processes. Recently, analyses of genome-wide chromatin packing in the model dicot plant Arabidopsis thaliana have been reported. At a kilobase scale, the A. thaliana chromatin interaction network is highly correlated with a range of genomic and epigenomic features. Surprisingly, topologically associated domains (TADs), which appear to be a prevalent structural feature of genome packing in many animal species, are not prominent in the A. thaliana genome. Using a genome-wide chromatin conformation capture approach, Hi-C , we report high-resolution chromatin packing patterns of another model plant, rice. We unveil new structural features of chromatin organization at both chromosomal and local levels compared to A. thaliana, with thousands of distinct TADs that cover about a quarter of the rice genome. The rice TAD boundaries are associated with euchromatic epigenetic marks and active gene expression, and enriched with a sequence motif that can be recognized by plant-specific TCP proteins. In addition, we report chromosome decondensation in rice seedlings undergoing cold stress, despite local chromatin packing patterns remaining largely unchanged. The substantial variation found already in a comparison of two plant species suggests that chromatin organization in plants might be more diverse than in multicellular animals.