Dynamics of A. thaliana/Pseudomonas pathosystem (bioRxiv)
Arabidopsis thaliana populations support long-term maintenance and parallel expansions of related Pseudomonas pathogens
With Kemen and Neher labs, we describe the natural dynamics of the A. thaliana/Pseudomonas pathsystem over several years and locations
Talia L Karasov, Juliana Almario, Claudia Friedemann, Wei Ding, Michael Giolai, Darren Heavens, Sonja Kersten, Derek S Lundberg, Manuela Neumann, Julian Regalado, Richard A Neher, Eric Kemen, Detlef Weigel
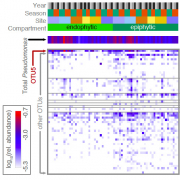
Crop disease outbreaks are commonly associated with the clonal expansion of single pathogenic lineages. To determine whether similar boom-and-bust scenarios hold for wild plant pathogens, we carried out a multi-year multi-site 16S rDNA survey of Pseudomonas in the natural host Arabidopsis thaliana. The most common Pseudomonas lineage corresponded to a pathogenic clade present in all sites. Sequencing of 1,524 Pseudomonas genomes revealed this lineage to have diversified approximately 300,000 years ago, containing dozens of genetically distinct pathogenic sublineages. The coexistence of diverse sublineages suggests that in contrast to crop systems, no single strain has been able to overtake these A. thaliana populations in the recent past. An important question for the future is how the maintenance of pathogenic diversity is influenced by host genetic variation versus an environment with more microbial and abiotic heterogeneity than in agricultural systems.