Talia’s paper on Pseudomonas population structure published
Arabidopsis thaliana and Pseudomonas Pathogens Exhibit Stable Associations over Evolutionary Timescales
OA | A single Pseudomonas lineage dominates local Arabidopsis populations
Karaso. Almario, Friedemann et al.
Cell Host & Microbe, Volume 24, Issue 1, 11 July 2018, Pages 155-167.e5
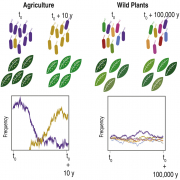
Crop disease outbreaks are often associated with clonal expansions of single pathogenic lineages. To determine whether similar boom-and-bust scenarios hold for wild pathosystems, we carried out a multi-year, multi-site survey of Pseudomonas in its natural host Arabidopsis thaliana. The most common Pseudomonas lineage corresponded to a ubiquitous pathogenic clade. Sequencing of 1,524 genomes revealed this lineage to have diversified approximately 300,000 years ago, containing dozens of genetically identifiable pathogenic sublineages. There is differentiation at the level of both gene content and disease phenotype, although the differentiation may not provide fitness advantages to specific sublineages. The coexistence of sublineages indicates that in contrast to crop systems, no single strain has been able to overtake the studied A. thaliana populations in the recent past. Our results suggest that selective pressures acting on a plant pathogen in wild hosts are likely to be much more complex than those in agricultural systems.