First preprint of 2019: The A. thaliana pan-NLRome
The Arabidopsis thaliana pan-NLRome
1000s of NLR genes in 65 diverse accessions
Van de Weyer et al., bioRxiv 537001, posted January 31, 2019
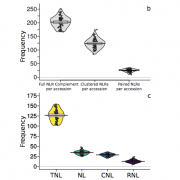
Disease is both among the most important selection pressures in nature and among the main causes of yield loss in agriculture. In plants, resistance to disease is often conferred by Nucleotide-binding Leucine-rich Repeat (NLR) proteins. These proteins function as intracellular immune receptors that recognize pathogen proteins and their effects on the plant. Consistent with evolutionarily dynamic interactions between plants and pathogens, NLRs are known to be encoded by one of the most variable gene families in plants, but the true extent of intraspecific NLR diversity has been unclear. Here, we define the majority of the Arabidopsis thaliana species-wide ‘NLRome’. From NLR sequence enrichment and long-read sequencing of 65 diverse A. thaliana accessions, we infer that the pan-NLRome saturates with approximately 40 accessions. Despite the high diversity of NLRs, half of the pan-NLRome is present in most accessions. We chart the architectural diversity of NLR proteins, identify novel architectures, and quantify the selective forces that act on specific NLRs, domains, and positions. Our study provides a blueprint for defining the pan-NLRome of plant species.