Cell-type specific RNA-seq and ChIP-seq
Phloem companion cell-specific transcriptomic and epigenomic analyses identify MRF1, a novel regulator of flowering
You et al. Plant Cell DOI: https://doi.org/10.1105/tpc.17.00714
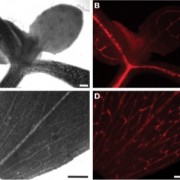
The phloem plays essential roles in source-to-sink relationship and in long-distance communication to coordinate growth and development throughout the plant. Here we employed INTACT coupled with low-input, high-throughput sequencing approaches to analyze the changes of the chromatin modifications H3K4me3 and H3K27me3 and their correlation with gene expression in the phloem companion cells (PCCs) of Arabidopsis thaliana shoots in response to changes in photoperiod. We observed a positive correlation between changes in expression and H3K4me3 levels of genes that are involved in essential PCC functions, including regulation of metabolism, circadian rhythm, development and epigenetic modifications. In contrast, changes in H3K27me3 signal appeared to contribute little to gene expression changes. These genomic data illustrate the complex gene-regulatory networks that integrate plant developmental and physiological processes in the PCCs. Emphasizing the importance of cell-specific analyses, we identified a previously uncharacterized MORN-motif repeat protein, MRF1, that was strongly upregulated in the PCCs in response to inductive photoperiod. mrf1 mutation delayed flowering whereas overexpression had the opposite effect, indicating that MRF1 acts as a floral promoter.