In Nature: collaborative paper on immunity
The EDS1–PAD4–ADR1 node mediates Arabidopsis pattern-triggered immunity
Rory N. Pruitt et al., Nature 598, 495–499 (2021)
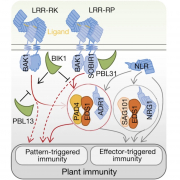
Plants deploy cell-surface and intracellular leucine rich-repeat domain (LRR) immune receptors to detect pathogens1. LRR receptor kinases and LRR receptor proteins at the plasma membrane recognize microorganism-derived molecules to elicit pattern-triggered immunity (PTI), whereas nucleotide-binding LRR proteins detect microbial effectors inside cells to confer effector-triggered immunity (ETI). Although PTI and ETI are initiated in different host cell compartments, they rely on the transcriptional activation of similar sets of genes2, suggesting pathway convergence upstream of nuclear events. Here we report that PTI triggered by the Arabidopsis LRR receptor protein RLP23 requires signalling-competent dimers of the lipase-like proteins EDS1 and PAD4, and of ADR1 family helper nucleotide-binding LRRs, which are all components of ETI. The cell-surface LRR receptor kinase SOBIR1 links RLP23 with EDS1, PAD4 and ADR1 proteins, suggesting the formation of supramolecular complexes containing PTI receptors and transducers at the inner side of the plasma membrane. We detected similar evolutionary patterns in LRR receptor protein and nucleotide-binding LRR genes across Arabidopsis accessions; overall higher levels of variation in LRR receptor proteins than in LRR receptor kinases are consistent with distinct roles of these two receptor families in plant immunity. We propose that the EDS1–PAD4–ADR1 node is a convergence point for defence signalling cascades, activated by both surface-resident and intracellular LRR receptors, in conferring pathogen immunity.

Our contribution: Extended Data Fig. 10: Classification of LRR-RPs, LRR-RKs and NLRs according to genetic conservation in Arabidopsis accessions.
a, Reads from 80 Arabidopsis accessions were mapped to the reference genome of Col-0. Genes were categorised as being conserved, having complex patterns of variation or exhibiting presence/absence polymorphisms according to the distribution of large-scale polymorphisms across all accessions as inferred from stringent read mappings. Criteria for categorization are detailed in the Methods. The numbers of genes falling into each category are provided in the corresponding bars. b, LRR-RP genes classified as in a. Genes encoding known immune receptors are indicated in bold.