Systematic transcriptomic study of heterosis
Pervasive under-dominance in gene expression as unifying principle of biomass heterosis in Arabidopsis
Yuan et al. (2022) bioRxiv 2022.03.03.482808
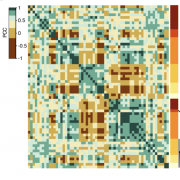
Heterosis, the generally superior performance in hybrids compared to their inbred parents, is one of the most enigmatic biological phenomena. Many different explanations have been put forward for heterosis, which begs the question whether common principles underpinning it do exist at all. We performed a systematic transcriptomic study in Arabidopsis thaliana involving 141 random crosses, to search for the general principles, if any, that heterotic hybrids share. Consistent additive expression in F1 hybrids was observed for only about 300 genes enriched for roles in stress response and cell death. Regulatory rare-allele burden affects the expression level of these genes but does not correlate with heterosis. Non-additive gene expression in F1 hybrids is much more common, with the vast majority of genes (over 90%) being expressed below parental average. These include genes that are quantitatively correlated with biomass accumulation in both parents and F1 hybrids, as well as genes strongly associated with heterosis. Unlike in the additive genes, regulatory rare allele burden in this non-additive gene set is strongly correlated with growth heterosis, even though it does not covary with the expression level of these genes. Together, our study suggests that while additive complementation is an intrinsic property of F1 hybrids, the major driver of growth in hybrids derives from the quantitative nature of non-additive gene expression, especially under-dominance and thus lower expression in hybrids than predicted from the parents.